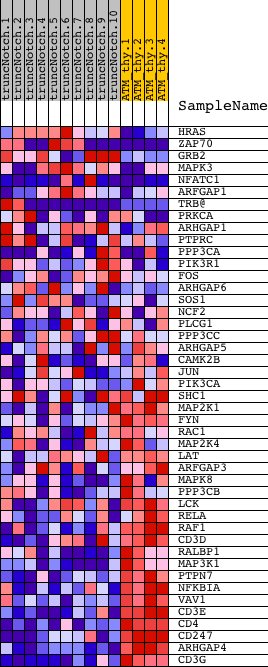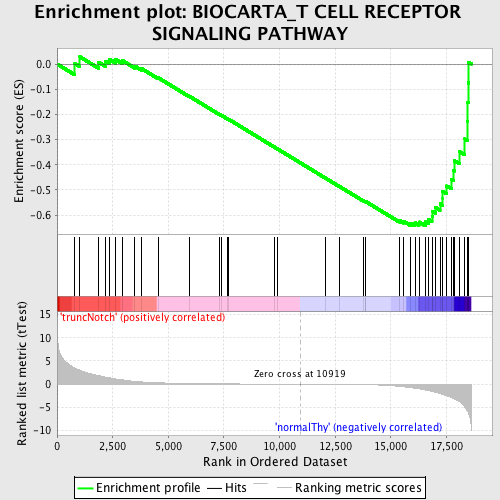
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy.phenotype_truncNotch_versus_normalThy.cls #truncNotch_versus_normalThy_repos |
| Phenotype | phenotype_truncNotch_versus_normalThy.cls#truncNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | -0.6428111 |
| Normalized Enrichment Score (NES) | -1.5287398 |
| Nominal p-value | 0.015706806 |
| FDR q-value | 0.37001345 |
| FWER p-Value | 0.993 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HRAS | 1980551 | 798 | 3.458 | 0.0019 | No | ||
| 2 | ZAP70 | 1410494 2260504 | 1024 | 2.988 | 0.0285 | No | ||
| 3 | GRB2 | 6650398 | 1856 | 1.865 | 0.0080 | No | ||
| 4 | MAPK3 | 580161 4780035 | 2169 | 1.520 | 0.0109 | No | ||
| 5 | NFATC1 | 510400 2320348 4050600 6180161 6290136 6620086 | 2365 | 1.357 | 0.0180 | No | ||
| 6 | ARFGAP1 | 4780364 5390358 | 2634 | 1.103 | 0.0179 | No | ||
| 7 | TRB@ | 3940292 | 2932 | 0.931 | 0.0140 | No | ||
| 8 | PRKCA | 6400551 | 3491 | 0.607 | -0.0082 | No | ||
| 9 | ARHGAP1 | 2810010 5270064 | 3803 | 0.490 | -0.0186 | No | ||
| 10 | PTPRC | 130402 5290148 | 4545 | 0.309 | -0.0545 | No | ||
| 11 | PPP3CA | 4760332 6760092 | 5932 | 0.142 | -0.1273 | No | ||
| 12 | PIK3R1 | 4730671 | 7309 | 0.074 | -0.2004 | No | ||
| 13 | FOS | 1850315 | 7375 | 0.071 | -0.2030 | No | ||
| 14 | ARHGAP6 | 2060121 | 7647 | 0.064 | -0.2168 | No | ||
| 15 | SOS1 | 7050338 | 7718 | 0.062 | -0.2197 | No | ||
| 16 | NCF2 | 540129 2370441 2650133 | 9783 | 0.019 | -0.3306 | No | ||
| 17 | PLCG1 | 6020369 | 9889 | 0.017 | -0.3361 | No | ||
| 18 | PPP3CC | 2450139 | 12057 | -0.021 | -0.4525 | No | ||
| 19 | ARHGAP5 | 2510619 3360035 | 12688 | -0.037 | -0.4859 | No | ||
| 20 | CAMK2B | 2760041 | 13753 | -0.088 | -0.5420 | No | ||
| 21 | JUN | 840170 | 13860 | -0.095 | -0.5465 | No | ||
| 22 | PIK3CA | 6220129 | 13874 | -0.096 | -0.5460 | No | ||
| 23 | SHC1 | 2900731 3170504 6520537 | 15383 | -0.470 | -0.6211 | No | ||
| 24 | MAP2K1 | 840739 | 15572 | -0.564 | -0.6239 | No | ||
| 25 | FYN | 2100468 4760520 4850687 | 15896 | -0.745 | -0.6316 | Yes | ||
| 26 | RAC1 | 4810687 | 16087 | -0.889 | -0.6303 | Yes | ||
| 27 | MAP2K4 | 5130133 | 16267 | -0.996 | -0.6270 | Yes | ||
| 28 | LAT | 3170025 | 16561 | -1.244 | -0.6267 | Yes | ||
| 29 | ARFGAP3 | 3390717 | 16691 | -1.364 | -0.6159 | Yes | ||
| 30 | MAPK8 | 2640195 | 16851 | -1.569 | -0.6041 | Yes | ||
| 31 | PPP3CB | 6020156 | 16865 | -1.582 | -0.5843 | Yes | ||
| 32 | LCK | 3360142 | 17003 | -1.730 | -0.5692 | Yes | ||
| 33 | RELA | 3830075 | 17212 | -1.973 | -0.5548 | Yes | ||
| 34 | RAF1 | 1770600 | 17308 | -2.124 | -0.5324 | Yes | ||
| 35 | CD3D | 2810739 | 17311 | -2.126 | -0.5049 | Yes | ||
| 36 | RALBP1 | 4780632 | 17514 | -2.449 | -0.4840 | Yes | ||
| 37 | MAP3K1 | 5360347 | 17735 | -2.897 | -0.4583 | Yes | ||
| 38 | PTPN7 | 3450110 | 17827 | -3.077 | -0.4233 | Yes | ||
| 39 | NFKBIA | 1570152 | 17856 | -3.162 | -0.3838 | Yes | ||
| 40 | VAV1 | 6020487 | 18073 | -3.720 | -0.3471 | Yes | ||
| 41 | CD3E | 3800056 | 18329 | -4.901 | -0.2973 | Yes | ||
| 42 | CD4 | 1090010 | 18448 | -5.828 | -0.2281 | Yes | ||
| 43 | CD247 | 3800725 5720136 | 18455 | -5.882 | -0.1521 | Yes | ||
| 44 | ARHGAP4 | 1940022 | 18482 | -6.085 | -0.0745 | Yes | ||
| 45 | CD3G | 2680288 | 18503 | -6.298 | 0.0061 | Yes |